Chronic Sinusitis & DNA Sequencing
Recently, I saw a young woman who had a two–year history of recurrent chronic rhinosinusitis (CRS). The patient’s symptoms began when she was transferred from a relatively new school (the patient is a school teacher) to a school that is over forty–years old. It was after the transfer that her symptoms worsened. In addition, there are apparently several other members of the faculty that are either experiencing sinus, or other respiratory problems. There is concern that there is a mold infestation in the school.
The patient was referred to me by an otolaryngologist (ENT) with whom we share several cases. He stated that the patient would have some improvement on oral antibiotics, but that all cultures showed no growth. He also stated that the patient was tried on itraconazole, an antifungal agent, on which she showed improvement, but developed an allergic reaction to the medication and it had to be stopped. Once the itraconazole was stopped, her symptoms returned.
The patient’s ENT had made the right assessment based on the historical evidence, mold infestation in the school, others with similar symptoms and clinical findings of both symptoms of congestion associated with the physical findings of inflamed mucosa and purulent discharge (pus). The frustrating part, not only in this case, is that it is an all–too–common scenario for those that treat patients with CRS.
Routine culture techniques are very unreliable for a number of different reasons, all from not being processed in a timely fashion to the fastidiousness of the pathological organisms. Thankfully there is another technique available known as DNA sequencing and this new technique has allowed many of us that do significant work in CRS management to prove what we have been saying for many years – that these chronic infections are polymicrobial (more than one organism) and that fungus may also be involved in the overall pathological process.
In another recent case, one of my long-term patients had been doing well on a nebulizer, using tobramycin (a broad-spectrum antibiotic) and steroids. However, when the patient became pregnant, the nebulizer was stopped. Though absorption of both medications is negligible when used topically, it was agreed between me and the patient to discontinue them so that no risk, no matter how small, was posed to the baby (who happens to be doing very well today).
Shortly after delivering the baby, the patient’s sinus condition returned. Routine culture grew MRSA, which the patient has had in the past. She was started on oral Doxycycline and had some improvement, but was still symptomatic. The patient told me later that her symptoms improved when she added oral Cipro to the regiment because she had some left over. The Cipro had no effect on the MRSA according to the sensitivities (antibiotics that work the best for that particular organism), yet the patient was feeling better. Why? Was this a placebo effect? Some would argue yes, but in both cases, DNA sequencing demonstrated what was truly going on with the patients.
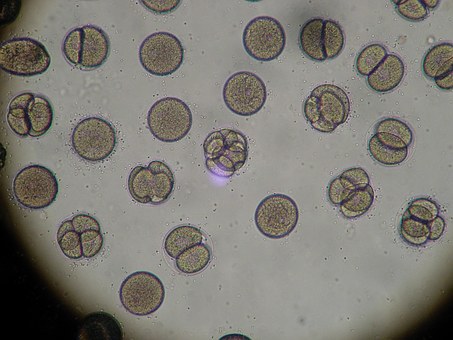
In the first case, using DNA sequencing, the patient was found to have three different types of mold in her sinuses including Alternaria, which has been associated with Allergic Fungal Sinusitis (AFS), though any type of fungus has the potential to cause AFS.
In the second case, the bacterial portion of MRSA was found to be only fifty percent of the pathological milieu causing the patient’s problem. In addition to MRSA, the patient was found to several species of Pseudomonas (all were sensitive to Cipro) and a species of Abiotrophia. Abiotrophia is a nasty little organism, which is usually involved with endocarditis (heart valve infections) and is also difficult to culture routinely. The concern with Abiotrophia is in its virulence, due to its ability to produce a large polysaccharide capsule, which not only protects the organism but contributes to the formation of biofilm (read my article on biofilm). Also in this second case, four different species of mold were identified – again including a species of Alternaria. In this second case, there is a clear demonstration of how AFS can lead to a secondary bacterial infection. Both cases are now on appropriate therapies and are doing well.
DNA sequencing works by amplifying the DNA of all the microbes found in a particular specimen. Once the DNA is amplified, it is then matched to a national database of all the microbes identified thus far, which is somewhere on the order of 13,000. Not only for me and my colleagues but for our patients, this new technology is allowing us to have a better understanding of the exact cause of a particular patient’s CRS and allows us to laser-guide our therapy to the individual. Hopefully, there will be an increased awareness of this technology which will help more sinus sufferers.